First Registration with DTI-TK
We assume that you have followed the Installation tutorial to download and install the DTI-TK binary packages.
Overview
This tutorial shows you how to register one DTI volume to another using DTI-TK. The goal is to demonstrate the registration performance of DTI-TK without you having to understand many intricacies of working with DTI volumes, discussed in depth in other tutorials. Hopefully, you will be satisfied with its performance and find it worthwhile to go on to understand how to properly prepare your dataset for use with DTI-TK!
In addition, it is very important that you run through this tutorial on your own computing environment. Reproducing the result of this tutorial is a good indicator that your system can handle DTI-TK correctly!
Download the example data set
The example data set contains the file "tensor.nii.gz", an example DTI volume in the DTI-TK compatible format, which we will use as the subject volume for registration.
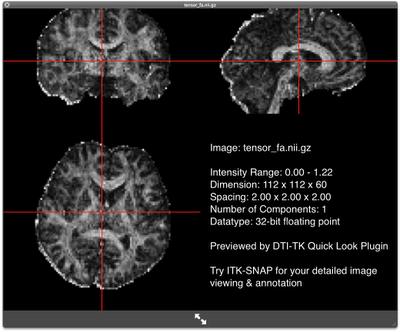 FA map of the subject FA map of the subjectDownload the IXI aging DTI template
The IXI aging DTI template contains, among other things, the file "ixi_aging_template.nii.gz", the template in the DTI-TK compatible format, which we will use as the template volume. We will also need the file "ixi_aging_template_brain_mask.nii.gz", which is a binary image specifying the area in the volume belonging to the brain parenchyma. The FA map of the template looks like this:
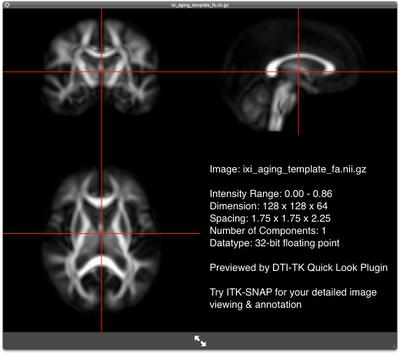 FA map of the template FA map of the templateOrganize the data for registration
For convenience, copy "tensor.nii.gz", "ixi_aging_template.nii.gz", and "ixi_aging_template_brain_mask.nii.gz" to a new directory of your choice.
Note: Make sure you are in the new directory before running the rest of the tutorial.
Run the rigid registration
The command to use:
dti_rigid_reg ixi_aging_template.nii.gz tensor.nii.gz EDS 4 4 4 0.01
The detailed usage of the command is given here. The output in the command window should look like this:
Rigid Registration of tensor.nii.gz to ../template/ixi_aging_template.nii.gz
Similarity Measure : EDS
Reorient Option : FS
Reading ../template/ixi_aging_template.nii.gz ... Done in 0.086205s
sampling step size = [2, 2, 2]
Gaussian smoothing: sigma = [0.873, 0.873, 0.624] ... Done in 0.333s
Reading tensor.nii.gz ... Done in 0.0609s
Gaussian smoothing: sigma = [0.736, 0.736, 0.736] ... Done in 0.188s
Initial Difference = 5.13e+06
initial estimate provided by center of mass alignment
cgm optimizer ftol = 0.01
both iteration 0 : 1.91e+06
both iteration 1 : 1.71e+06
both iteration 2 : 1.66e+06
both iteration 3 : 1.64e+06
cpu time consumed in seconds: 1.4
Final Registration Parameters = 106 -58.1 -60.9 0.158 -0.033 -0.108
Final Difference = 1.64e+06
Total Difference Computation = 30
Total Gradient Computation = 4
Output the rigid transformation as tensor.aff
inFile = tensor.nii.gz
outFile = tensor_aff.nii.gz
interpolation option is LEI
reorientOption = PPD
Reading tensor.nii.gz ... Done in 0.068549s
trans (inverse) applied = [120.391, -79.026, -43.910]
[ 0.994, -0.102, -0.033; 0.106, 0.982, 0.158; 0.016, -0.160, 0.987]
reading output volume specification from ../template/ixi_aging_template.nii.gz
backward resampling ...time consumed = 0.431
Writing tensor_aff.nii.gz ... Done in 0.426s
done
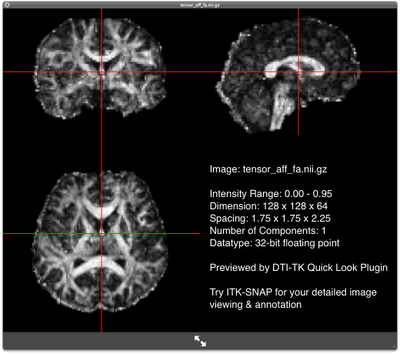 FA map of the rigidly-aligned subject FA map of the rigidly-aligned subjectOn a 3GHz Intel core 2 duo machine, the program takes less than a minute to run. It will create two new files "tensor_aff.nii.gz", the rigidly aligned DTI volume, and "tensor.aff", the corresponding rigid transformation.
Run the affine registration
The command to use:
dti_affine_reg ixi_aging_template.nii.gz tensor.nii.gz EDS 4 4 4 0.01 1
The detailed usage of the command is not given here, since the output in the command window looks very similar to that of the rigid registration. The program takes about a minute to run and will overwrite the "tensor_aff.nii.gz" and "tensor.aff" with the newer version, i.e., the affine aligned volume and the corresponding affine transformation.
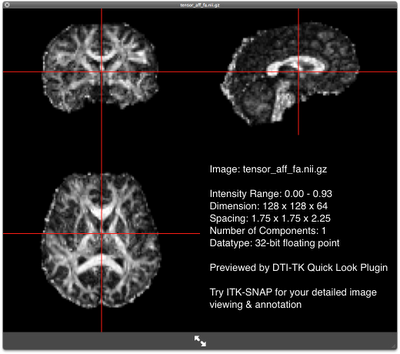 FA map of the affinely-aligned subject FA map of the affinely-aligned subjectRun the diffeomorphic registration
The command to use:
dti_diffeomorphic_reg ixi_aging_template.nii.gz tensor_aff.nii.gz ixi_aging_template_brain_mask.nii.gz 1 5 0.002
The detailed usage of the command is given here. It registers the subject to the template in an iterative process. For one iteration, the output in the command window looks like this:
registering tensor_aff.nii.gz to ../template/ixi_aging_template.nii.gz ...
starting at Tue 14 May 2013 15:17:12 BST
iteration 1 begins ...
Similarity Measure : DDS, Reorient Option : FS
Reading ../template/ixi_aging_template.nii.gz ... Done in 0.089588s
Reading tensor_aff_diffeo.nii.gz ... Done in 0.07375s
Reading ../template/ixi_aging_template_brain_mask.nii.gz ... Done in 0.0027s
starting level 2, ending level 5
Level 2
Piecewise Affine Setup: size = 8, 8, 4; vsize = 28, 28, 36
pDim = 3072; scalings : img = 0.06, prior = 0.1, reg = 0.04; ftol = 0.002
before : sum = 8158.72, img = 135979, prior = 0, reg = 0
iter 0: sum = 8158.72, img = 135979, prior = 0, reg = 0
iter 1: sum = 7210.51, img = 112827, prior = 85.6186, reg = 10808.2
iter 2: sum = 7062.5, img = 115089, prior = 101.244, reg = 3675.72
iter 3: sum = 6969.66, img = 114319, prior = 214.507, reg = 2226.6
iter 4: sum = 6790.31, img = 108454, prior = 610.284, reg = 5551.64
iter 5: sum = 6383.92, img = 97723.5, prior = 1522.14, reg = 9207.51
iter 6: sum = 6170.33, img = 94188.1, prior = 1652.23, reg = 8845.62
iter 7: sum = 6064.83, img = 92752.7, prior = 1650.75, reg = 8364.8
iter 8: sum = 5976.85, img = 91041, prior = 1612.5, reg = 8828.51
iter 9: sum = 5878.97, img = 90597.4, prior = 1507.46, reg = 7309.53
iter 10: sum = 5833.3, img = 91212.9, prior = 1463.47, reg = 5354.62
iter 11: sum = 5821.98, img = 91344.5, prior = 1452.53, reg = 4901.4
iter = 79, iterGrad = 12
after : sum = 5816.1, img = 91272.4, prior = 1462.49, reg = 4838.15
Level 3
Piecewise Affine Setup: size = 16, 16, 8; vsize = 14, 14, 18
pDim = 24576; scalings : img = 0.06, prior = 0.1, reg = 0.04; ftol = 0.002
before : sum = 5816.1, img = 91268.9, prior = 1463.12, reg = 4841.4
iter 0: sum = 5816.1, img = 91268.9, prior = 1463.12, reg = 4841.4
iter 1: sum = 5625.25, img = 88799.8, prior = 1405.19, reg = 3918.67
iter 2: sum = 5559.75, img = 88363.7, prior = 1443.22, reg = 2840.02
iter 3: sum = 5480.6, img = 85541.3, prior = 1714.91, reg = 4415.75
iter 4: sum = 5435.87, img = 83679.9, prior = 2009.07, reg = 5354.16
iter 5: sum = 5401.83, img = 82336.5, prior = 2364.7, reg = 5629.19
iter 6: sum = 5382.33, img = 81653.4, prior = 2578.97, reg = 5630.57
iter 7: sum = 5369.56, img = 81257.3, prior = 2703.85, reg = 5593.42
iter 8: sum = 5354.88, img = 80747.9, prior = 2830.33, reg = 5674.36
iter 9: sum = 5339.62, img = 79964.5, prior = 3006.74, reg = 6026.97
iter 10: sum = 5321.12, img = 78827.7, prior = 3314.55, reg = 6500.24
iter 11: sum = 5306.74, img = 78208.9, prior = 3496.09, reg = 6614.74
iter 12: sum = 5291.45, img = 77610.8, prior = 3630.61, reg = 6793.49
iter 13: sum = 5278.73, img = 77181, prior = 3726.12, reg = 6881.43
iter 14: sum = 5269.66, img = 77015.4, prior = 3759.82, reg = 6818.98
iter = 118, iterGrad = 15
after : sum = 5265.43, img = 77061.1, prior = 3728.67, reg = 6722.72
Level 4
Piecewise Affine Setup: size = 32, 32, 16; vsize = 7, 7, 9
pDim = 196608; scalings : img = 0.06, prior = 0.1, reg = 0.04; ftol = 0.002
before : sum = 5265.43, img = 77063.6, prior = 3727.06, reg = 6722.73
iter 0: sum = 5265.43, img = 77063.6, prior = 3727.06, reg = 6722.73
iter 1: sum = 4993.26, img = 74081.1, prior = 3296.8, reg = 5467.9
iter 2: sum = 4926.22, img = 73720.4, prior = 3224.65, reg = 4513.37
iter 3: sum = 4863.62, img = 71868.2, prior = 3215.34, reg = 5749.89
iter 4: sum = 4836.11, img = 70807.6, prior = 3314.56, reg = 6404.92
iter 5: sum = 4812.95, img = 69964.3, prior = 3556.42, reg = 6486.28
iter 6: sum = 4800.12, img = 69570.3, prior = 3729.15, reg = 6324.78
iter 7: sum = 4793.56, img = 69422.8, prior = 3796.2, reg = 6214.18
iter = 65, iterGrad = 8
after : sum = 4790.48, img = 69411.8, prior = 3774.2, reg = 6209.05
Level 5
Piecewise Affine Setup: size = 64, 64, 32; vsize = 3.5, 3.5, 4.5
pDim = 1572864; scalings : img = 0.06, prior = 0.1, reg = 0.04; ftol = 0.002
before : sum = 4790.48, img = 69411.4, prior = 3773.09, reg = 6212.3
iter 0: sum = 4790.48, img = 69411.4, prior = 3773.09, reg = 6212.3
iter 1: sum = 4581.37, img = 66837.3, prior = 3473.34, reg = 5595.1
iter 2: sum = 4523.92, img = 66511, prior = 3310.61, reg = 5055.02
iter 3: sum = 4428.27, img = 63992.3, prior = 2864.49, reg = 7557.09
iter 4: sum = 4380.25, img = 62494.1, prior = 2716.68, reg = 8973.4
iter 5: sum = 4364.7, img = 62142.7, prior = 2769.34, reg = 8980.19
iter 6: sum = 4347.86, img = 61840.1, prior = 2919.03, reg = 8638.8
iter 7: sum = 4339.33, img = 61704.8, prior = 3034.09, reg = 8340.82
iter = 65, iterGrad = 8
after : sum = 4335.43, img = 61614.8, prior = 3042.83, reg = 8356.92
Little Endian System
Writing VECTORS ...
size is currently determined internally.
vsize = 1, 1, 1
origin is currently default to [0, 0, 0].
outFile = tensor_aff_to_ixi_aging_template.5.df.nii.gz
Little Endian System
Reading VECTORS ...
Writing tensor_aff_to_ixi_aging_template.5.df.nii.gz ... Done in 4.16411s
converting to the diffeomorphic deformation field: ...
Reading tensor_aff_to_ixi_aging_template.5.df.nii.gz ... Done in 0.520504s
Gaussian smoothing: sigma = [ 1, 1, 1] ... Done in 1.77s
Gaussian smoothing: sigma = [ 1, 1, 1] ... Done in 1.7s
maxNorm = 9.49 iterations = 18
Voxelwise scaling tensor_aff_to_ixi_aging_template.5.df.nii.gz by 3.81e-06 ... Done in 0.0356s
. . . . . . . . . . . . . . . . . .
Writing tensor_aff_to_ixi_aging_template.5.df.nii.gz ... Done in 4.09s
Reading ../template/ixi_aging_template_brain_mask.nii.gz ... Done in 0.002721s
input volume ../template/ixi_aging_template_brain_mask.nii.gz
size: 128x128x64, voxel size: 1.75x1.75x2.25, origin: [0, 0, -0]
reading output volume specification from tensor_aff_to_ixi_aging_template.5.df.nii.gz
output volume specification: size: 224x224x144, voxel size: 1x1x1, origin: [0, 0, 0]
matching the center of the old and the new voxel spaces
Writing tensor_aff_jac_mask.nii.gz ... Done in 0.259388s
inFile = tensor_aff_to_ixi_aging_template.5.df.nii.gz
Reading tensor_aff_to_ixi_aging_template.5.df.nii.gz ... Done in 0.518063s
outFile = tensor_aff_to_ixi_aging_template.5.df_jac.nii.gz
Writing tensor_aff_to_ixi_aging_template.5.df_jac.nii.gz ... Done in 2.87775s
JACOBIAN STATISTICS: after current iteration mean = 0.980048 min = 0.487728 max = 1.87831 # of voxels = 1.78546e+06
inFile = tensor_aff.nii.gz
outFile = tensor_aff_diffeo_current.nii.gz
transFile = tensor_aff_to_ixi_aging_template.5.df.nii.gz
df option is FD
interpolation option is LEI
reorientOption = PPD
Reading tensor_aff.nii.gz ... Done in 0.079715s
Reading tensor_aff_to_ixi_aging_template.5.df.nii.gz ... Done in 0.531295s
backward resampling ...time consumed = 2.34449
Writing tensor_aff_diffeo_current.nii.gz ... Done in 0.667442s
IMAGE SIMILARITY: after previous iteration = 135992 after current iteration = 71756
iteration 1 done
The program takes about 5 minutes or less to complete for each iteration. The number of iterations is an input argument that can be adjusted for your need. The example given uses 5 iterations and completes in less than 20 minutes. The program will generate a pair of new files that include: "tensor_aff_diffeo.nii.gz", the diffeomorphic aligned volume, "tensor_aff_diffeo.df.nii.gz", the corresponding deformation field. There are a few other intermediate files will be created during the registration but will be removed when the registration completes.
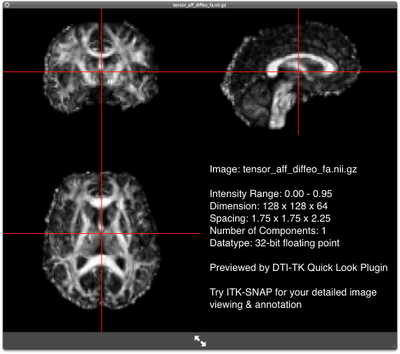 FA map of the deformably-aligned subject FA map of the deformably-aligned subjectWhat's Next?
If you are now convinced that you should give DTI-TK a try on your own data set, you should proceed by
- Reading the tutorials on data format, visualization and checking diffusivity unit
- Going through the detailed preprocessing tutorial
- Studying the detailed registration tutorial.
|
|